Life sciences and healthcare are now in the limelight for technologists, in part because AI technologies are well suited to make a positive impact to key workflows. In this essay, I'll explore 6 areas of life sciences that offer fruitful applications of AI. I hope this will serve as a resource and point of inspiration for those of you who are interested to work in this field.
As usual, just hit reply if you'd like to share thoughts/critique/your work! We'll resume our regular market coverage in the next issue. -Nathan
🔬 Setting the scene
In 2013, the machine learning (ML) research community demonstrated the uncanny ability for deep neural networks trained with backpropagation on graphics processing units to solve complex computer vision tasks. The same year, I wrapped up my PhD in cancer research that investigated the genetic regulatory circuitry of cancer metastasis. Over the 6 years that followed, I’ve noticed more and more computer scientists (we call them bioinformaticians :) and software engineers move into the life sciences. This influx is both natural and extremely welcome. Why? The life sciences have become increasingly quantitative disciplines thanks to high-throughput omics assays such as sequencing and high-content screening assays such as multi-spectral, time-series microscopy. If we are to achieve a step-change in experimental productivity and discovery in life sciences, I think it’s uncontroversial to posit that we desperately need software-augmented workflows. This is the era of empirical computation (more on that here). But what life science problems should we tackle and what software approaches should we develop?
AI methods in life sciences
I’ve maintained a subscription to Nature Methods ever since my senior year in undergrad. Looking at the cover of each issue gives me a temperature check for what scientists think is new and exciting today. Over the last 12 months, there’s seldom been an issue that runs without a feature story about a new ML solution to a life science problem. These tasks tend to have previously been “solved” by hand or with a brittle, rules-based system from the 90s. Below you’ll find a snapshot of the 2018 Nature Methods covers. Can you see a trend?
The papers making the headlines include 3D super-resolution microscopy, DeepSequence for variant calling, 3D cell segmentation and tracking animal behaviour through video analysis. These tasks involve high-dimensional input data, an agreed taxonomy of labels, and large volumes of data that is becoming cheaper to produce at scale. As such, ML methods can likely help classify, cluster and generate novel data points for these tasks. If we sample Nature, Nature Methods, Nature Biotechnology and Nature Medicine papers over the last 5 years, we can can see that in fact over 80% of papers mentioning “artificial intelligence” or “deep learning” were published in the last 2 years alone!
6 impactful applications of AI to the life sciences
Two years ago I wrote a piece describing 6 areas of machine learning to watch closely. In this post, I’ll describe 6 areas of life science research where AI methods are making an impact. I describe what they are, why they are important, and how they might be applied in the real world (i.e. outside of academia or industrial research groups). I’ll start from the small scale (molecules) and work up to the larger scale (cells). I look forward to hearing your comments and critique 👉@nathanbenaich.
1. Molecular property prediction
Drug discovery can be viewed as a multi-parameter optimisation problem that stretches over vast length scales. Successful drugs are those that exhibit desirable molecular, pharmacokinetic and target binding properties. These pharmacokinetic and pharmacology properties are expressed as absorption, distribution, metabolism, and excretion (ADME), as well as toxicity in humans and protein-ligand (i.e. drug to protein target) binding affinity. Traditionally, these features are examined empirically in vitro using chemical assays and in vivo using animal models. To do so, most academic labs will rely on lab scientists endlessly pipetting and transferring small amounts of liquids between plastic vials, tissue culture and various pieces of analytical equipment. Pharmaceutical companies will do much of the same, but they might have budget for purchasing robotic automation equipment for tissue culture and liquid handling.
There are also computational methods that help elucidate molecular properties in silico. However, most computational efforts have been based in some way on physics, i.e., calculating energies from quantum mechanics or doing thermodynamics-based simulations, or simple empiricism. In contrast to ML-based approaches that will learn to approximate the underlying mechanisms of a target process from raw data, these physics-based computational tools will only capture the underlying mechanisms that developers have explicitly encoded during the development process. Thus, from an ML point of view, physics-based computational tools are essentially feature-engineered and thus rather brittle and do not generalise well to out of sample distributions.
Over the last two years or so, the ML research community has produced a flurry of papers describing novel deep learning methods to predict the properties of small molecule drugs and proteins therapeutics. A driver of success or failure for these ML approaches rests in the computational representations that these models use to learn the intrinsic rules of chemistry that are encoded in raw experimental data. Broadly speaking, there are three representation types for chemical molecules that are used today: Extended Connectivity Fingerprint (ECFP, a fixed length bit representation), Simplified Molecular Input Line Entry System (SMILES, where molecules are essentially represented as a linear string of letters, or DeepSMILES, an updated syntax that solves invalid syntax issues when training probabilistic models), and graph-based representations (where nodes are atoms and edges are the chemical bonds linking atoms). Each representation imposes different constraints on model performance and a machine’s ability to understand the data. Here, ECFP is extracted from SMILES, and SMILES is extracted from a 3D conformations of the molecule graph. Each transformation process destroys a certain amount of data as a trade for machine readability and computability. The following figure illustrates this concept. The ideal scenario is to use the most comprehensive molecular representation that a machine can fully utilise.
A first set of notable papers exploit the fact that molecules can be efficiently represented as molecular graphs. These include systems such as PotentialNet from Stanford, Message Passing Neural Networks from GoogleAI, Graph Memory Networks from Deakin University as well as benchmarks such as MoleculeNet from Stanford. The second set of papers use more simplistic representations such as SMILES, molecular fingerprints, or N-gram graphs. These include systems such as CheMixNet from Northwestern, N-Gram Graph from Wisconsin-Madison, Multi-Modal Attention-based Neural Networks from IBM Research and ETH Zurich.
The appeal of these deep learning systems is that they can automatically learn the underlying rules that relate molecular composition and structure to chemical properties from raw experimental data. Indeed, the ML models mentioned here can predict >10 chemical properties accurately enough to be useful to chemists. Most importantly, these ML approaches can generate property predictions up to 300,000 times faster than it would take to simulate them using computationally-intensive numerical approximations to Schrödinger’s equation. This means that a chemist might no longer have to run a flurry of chemical assays to empirically determine a molecule’s chemical properties. She can instead draw a molecule in ChemDraw and an ML model will immediately return its chemical properties! One could imagine using molecular property predictions as a means of scoring large-scale virtual drug library screens. Here, a chemist would explicitly stipulate her desired properties and a screening system would score a large library of molecules against these properties to return the best fitting candidates. Going further, we could even consider exploring the search space of yet-to-be-synthesised compounds to more rationally direct development efforts. This is all made possible because ML approaches to computational chemistry learn to approximate the underlying mechanisms of chemistry after training on datasets of molecules and property pairings.
2. Generating novel chemical structures
There are an estimated 1,453 drugs that have been approved by the US Food and Drug Administration (FDA) in its entire history. If we take small molecules as a case study (as opposed to protein therapeutics), most drug libraries that can be purchased from suppliers come in batches of 1,000 different molecules. This is literally a drop in the ocean because computational estimates suggest that the actual number of small molecules could be 10^60 (i.e. 1 million billion billion billion billion billion billion). This is 10-20 orders of magnitude less some estimates of the number of atoms in the universe (for readers who are familiar with the AlphaGo story, this comparison may sound familiar). The American Chemical Society Chemical Abstracts Service database contains almost 146 million organic and inorganic substances, which suggests that almost all small molecules (>99.9%) have never been synthesized, studied and tested.
Taken together, these statistics mean that research labs can only test a few thousand drug candidates in the real world. Once they’ve found a few candidates that look functionally promising, they often iteratively explore structural variants to determine if there are better versions out there. This again requires synthesis experiments and chemical assays. As such, to even make a dent into the 10^60 drug-like small molecules in existence, we need to use software as a means to rationally and automatically explore the chemical design space for candidates that have desired properties for a specific indication.
To solve this problem, ML researches have turned to sequence-based generative models trained on large databases of chemical structures (often using SMILES representations). Here, chemistry is effectively treated as a language with its own grammar and rule set. Generative models are capable of learning the probability distribution over molecule space such that they implicitly internalise the rules of chemistry. This compares somewhat to how generative models trained over a corpus of English text can learn English grammar. To this end, a 2017 paper from the University of Münster and AstraZeneca presented a recurrent neural network that was trained as a generative model for molecular structures. While the performance wasn’t state-of-the-art, the system could reproduce a small percentage of molecules known to have antibacterial effects. Related work from AstraZeneca expanded on the RNN-based molecule generator by using a policy-based reinforcement learning approach to tune the RNN for the task of generating molecules with predetermined desirable properties. The authors show that the model can be trained to generate analogues to the anti-inflammatory drug Celecoxib, implying that this system could be used for scaffold hopping or library expansion starting from a single molecule.
Other ML methods have been used to build molecule generators. The University of Tokyo and RIKEN borrowed from the AlphaGo architecture to build a system called ChemTS. Here, a SMILES-based molecule generator system is built using Monte Carlo Tree Search for shallow search and an RNN for rollout of each downstream path. The same group also published an evolutionary approach called ChemGE that relies on genetic algorithms to improve the diversity of molecules that are ultimately generated. Finally, researchers at the University of Cambridge applied a pair of deep networks trained as an autoencoder to convert molecules represented as SMILES strings into a continuous vector representation. The autoencoder was trained jointly on a property prediction task. Finally, even generative adversarial networks (GANs) have been adapted to operate directly on graph-structure molecule data. The MolGAN system from the University of Amsterdam combines GANs with a reinforcement learning objective to encourage the generation of molecules with specific desired chemical properties. Taken together, these papers illustrate the promise for accelerating chemical search space exploration using virtual screening.
3. Chemical synthesis planning
Now that we’ve discussed AI methods for exploring the chemical design space and predicting pharmaceutically-relevant chemical properties of these molecules, the next problem for AI to tackle would be chemical synthesis planning. Chemists use retrosynthesis to figure out the most efficient stepwise chemical reaction process to produce a small molecule of interest. This task is similar (albeit admittedly more complex) to being given a cake and being asked to design the optimal ingredient list and recipe to recapitulate the same cake by working backwards. Given that we’re talking about chemical reactions, the transformations from one molecule state to another are derived from a database of successfully conducted similar reactions with analogous starting materials. These reactions often carry the name of their scientific discoverers. Chemists intuitively select from these reactions, mix and match, and empirically assess the efficiency with which each reaction generates sufficient yield of a desired product. If a molecule doesn’t react as predicted it is “out of scope”. If the molecule reacts as predicted it is “in scope”. Predicting which will occur is challenging and uncertain.
To address this task, researchers at the University of Münster and Shanghai University developed a system for rapid chemical synthesis planning. The system uses three neural networks and Monte Carlo Tree Search (together called 3N-MCTS). It was trained on 12.4 million single-step reactions, which is essentially all reactions ever published in the history of organic chemistry. Paraphrasing from their introduction, “The first neural network (the expansion policy) guides the search in promising directions by proposing a restricted number of automatically extracted transformations. A second neural network then predicts whether the proposed reactions are actually feasible (in scope). Finally, to estimate the position value, transformations are sampled from a third neural network during the rollout phase.”
What’s interesting is that their method is far faster that the state-of-the-art computer-assisted synthesis planning. In fact, 3N-MCTS solves more than 80% of a molecule test set with a time limit of 5 seconds per target molecule. By contrast, an approach called best first search in which functions are learned through a neural network can solve 40% of the test set. Best first search designed with hand-coded heuristic functions performs the worst: it solves 0% in 5s. If provided with infinite runtime, however, the three approaches will converge to the same performance. One could imagine that automated chemical synthesis planning methods like 3N-MCTS should tie in with virtual screening approaches to remove some heavy lifting from chemists.
4. Protein structure prediction
When we think about health and disease, we often focus on genetics. However, the manifestation of phenotypes is actually exerted by proteins, which are encoded by our genetics. For example, mutated genes might encode erroneous proteins that behave the wrong way in pathways responsible for cell division, which could drive the onset of cancer. Moreover, the drugs we take to treat health conditions act by binding to other proteins or DNA in order to modulate the cellular processes that underpin that condition. What’s more, we live in a three dimensional world (with time as a fourth dimension). This means that understanding how proteins work and how they contribute to health and disease is made a lot easier by having their 3D structure, not just their amino acid sequence.
The problem, however, is that generating a 3D structure for a protein is expensive, complicated and time-consuming. The process requires a protein of interest to be isolated, purified, crystallised and X-Rays blasted at the protein to measure how the beams diffract off the crystal surface. As a result, there are orders of magnitude more proteins in the human body than crystal structures available in public databases. The number of complete amino acid sequences also outnumbers the number of crystal structures available in public databases by orders of magnitude. Researchers have therefore been working on computationally predicting the 3D structure of a protein from its amino acid sequence for years. This is the “protein folding problem”.
Recently, DeepMind published a system called AlphaFold that uses two deep residual convolutional neural networks (CNNs) to reach a new state-of-the-art for protein folding prediction. Specifically, the system predicts (a) the distances between pairs of amino acids and (b) the angles between chemical bonds that connect those amino acids. By predicting how close pairs of amino acids are to one another, the system creates a distance map of the protein. This map can essentially be extrapolated to generate a 3D structure or match one from an existing database. For a longer critical review of the work that is outside the scope of this piece, please check out this post. In theory, AlphaFold could enable scientists to generate de novo 3D protein structures for novel proteins that they discover or for known proteins that have yet to be crystallised.
Furthermore, one can imagine AlphaFold being integrated into drug development pipelines as an additional signal to predict whether a drug of interest will indeed bind its target and, if so, with what affinity.
More work is needed to expand AlphaFold to predict the 3D structure of protein complexes (i.e. several individual proteins working together) and to make the system’s predictions more dependable. On the former point, recent research from Stanford presented the Siamese Atomic Surfacelet Network could help. Their system is an end-to-end learning method for protein interface prediction that uses representations learned from raw spatial coordinates and identities of atoms as inputs. This network predicts which surfaces of one protein bind to which surfaces of another protein. It is worth noting that the 3D conformation of proteins before, during and after binding can change significantly and are also affected by their environment. Models like this one will need to be hugely flexible to accomodate for such real-world nuances.
5. Protein identification using mass spectrometry
In order to elucidate the drivers of health and disease, we must discern which genes and proteins are functionally connected to a phenotype of interest. To do so, we might design experiments where we induce a disease phenotype and compare the genomic and proteomic profiles of diseased cells with healthy cells. If we focus on the proteome, a key method for identifying and quantifying the proteins, their complexes, subunits and functional interactions that are present in a cell mixture is called protein mass spectrometry. Briefly, this protocol works by taking a protein mixture sample, separating the constituent peptides (short sequences of amino acids), ionising them and separating them on a spectrum according to their abundance and mass/charge ratios. The result is a histogram (called a chromatogram) of the different ways that a peptide can fragment where each fragment appears as a peak on the plot. A scientist must interpret this pattern of peaks to find fingerprints that are unique to specific peptides that make up a whole protein. Only by doing so can they reconstruct the protein population in the sample mixture.
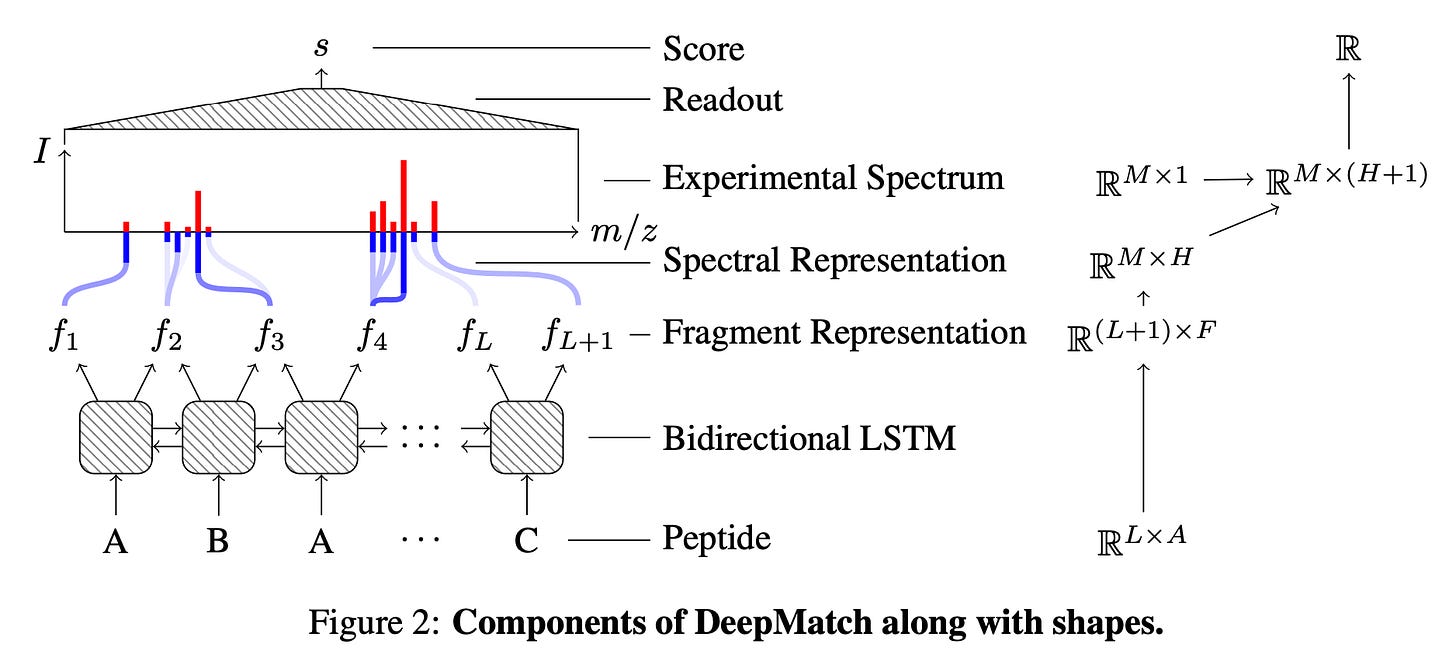
While there are a variety of open source and proprietary software tools to analyse mass spectrometry data, a central challenge to data interpretation is that there are few ground truth pairings between peptides and spectra. This means that ML methods in this domain must be able to use existing noisy labels to make progress. Recent work from GoogleAI, Calico Life Sciences and InSitro present a deep learning system called DeepMatch (schematic below) that can identify peptides corresponding to a given mass spectrum based on noisy labels from classical algorithms. The model is also trained to distinguish between genuine true spectra and dummy synthetic negative spectra to help with overcoming overfitting. DeepMatch achieves a 43% improvement over standard matching methods and a 10% improvement over a combination of the matching method and an industry standard cross-spectra reranking tool.
One can imagine that techniques like DeepMatch form part of the early phases of drug discovery where scientists need to rapidly, scalably and more reliably quantify protein candidates that have a potentially causative relationship with a disease phenotype of interest. Interestingly, ML methods applied to mass spectrometry can also be used to for chemicals where the task is to predict the presence of substructures given the mass spectrum, and matching these substructures to the candidate structures.
6. Automated analysis of experimental image data
While virtual screening is very appealing, the power of these computational models depends on three key factors. These include the model architecture, the data representation it uses, and the availability of high-quality ground truth data. For empirical computation to deliver on its promise, we must close the computational-to-real-world feedback loop with rationally-guided wet lab experiments such as omics sequencing and high-content microscopy. These results serve as our ground truth. However, as the volumes, dimensions and resolution of ground truth and experimental datasets grow, the methods we use for data analysis are also due for a refresh with more flexible and reliable ML-driven replacements.
Let’s consider high-throughput drug screens using human cancer cells cultured in vitro as our model system. Assume that we are running a hypothesis-generating experiment whereby many single drug candidates and combinations thereof are tested for their ability to halt the division of human breast cancer cells. In order to detect cancer cells that no longer divide, we must detect and quantify markers of this cell state. We might be interested in localising the cell’s nucleus, its cytoskeleton, as well as proteins that regulate cell division and mark the stage of its cell cycle. The workhorse method we use involves binding a fluorescent antibody probe to the protein marker of choice such that we can localise and quantify the extent of its presence or absence within a cell by fluorescent microscopy. This is called immunofluorescence. There are several technical and workflow challenges to this protocol. For example, sample preparation and staining is quite laborious, results are sensitive to concentrations of target and probe, the protocol is rather inconsistent and is terminal in the sense that the sample cannot be reused afterwards. We’re also limited in the number of proteins we can visualise because of overlap on the fluorescence spectrum. Finally, we’re only “lighting up” what we believe to be important for analytical purposes. Cells are incredibly complex organisms and we might very well be missing key features that help us understand their biology.
Tying back to the opening of this piece, computer vision has come leaps and bounds for complex feature learning in 2D and 3D images thanks to deep learning. This means that we should be able to apply deep learning to automate the analysis of high-content microscopy images. Last year, a collaboration led by UCSF and Google resulted in a deep learning system capable of reliably predicting several fluorescent labels (nuclei, cell type, cell state) from unlabeled transmitted light microscopy images of biological samples. The method is consistent because it is computational and is not limited by spectral overlap. More importantly, in silico predictions mean that we no longer need to “waste” the sample on immunofluorescence. The trained network exhibits transfer learning too, which means that once it has learned to predict a set of labels, it can learn new labels with a small number of additional data.
While this study predicted immunofluorescence in 2 dimensions, a study from the Allen Institute in Seattle demonstrated a system to do the same but in 3 dimensions. This method can therefore generate multi-structure, integrated images and can also be extended to predict immunofluorescence from electron micrographs (more detailed, higher magnification images of smaller cellular structures). The Allen Institute paper used a U-Net CNN architecture developed by Olaf Ronnenberger. Indeed, Olaf’s group recently applied the U-Net for automated cell counting, detection and morphometry that comes as a simple plugin to the popular (but very basic) image analysis software tool called ImageJ. Side joke: I always had the feeling ImageJ was like Microsoft Paint for biologists 😂. The U-Net was also used by DeepMind and Moorfield’s Eye Hospital in their Nature Medicine paper for segmenting optical computed tomography scans of the eye. Finally, GANs have proven useful for transforming diffraction-limited input images into super-resolved ones, which could one day obviate the need for using expensive super resolution microscopes. Indeed, a study from UCLA developed a framework to improve the resolution of wide-field images acquired with low-numerical-aperture objectives, matching the resolution that is acquired using high-numerical-aperture objectives. Neat! Taken together, it is clear that computer vision methods applied to microscopy will enable much more reliable and automated visual data interpretation. Doing so could potentially open new questions and offer new takes on our current understanding of biology.
Conclusion
I hope that this essay has shined light onto current progress around AI applications to life sciences and pointed to the immense potential in front of us. This piece is by no means comprehensive and somewhat reflects my personal bias of what I find impactful and scientifically interesting. If you’d like me to include or feature your work, don’t hesitate to get in touch in the comments or drop me a note 👉@nathanbenaich.
---
Signing off,
Nathan Benaich, 24 February 2019
Air Street Capital | Twitter | LinkedIn | RAAIS | London.AI
Air Street Capital is a venture capital firm that invests in AI-first technology and life science companies. We’re a team of experienced investors, engineering leaders, entrepreneurs and AI researchers from the World’s most innovative technology companies and research institutions.