Design of highly functional genome editors by modelling CRISPR–Cas sequences
Today marks a significant scientific milestone for frontier AI systems in biology. Our portfolio company Profluent has its work on OpenCRISPR-1, the first AI-designed CRISPR-Cas protein that can precisely edit human DNA, published in the highest impact scientific journal, Nature.
This work demonstrates how generative language models can design functional, novel genome editors from scratch that outperform natural and previously engineered systems on key criteria including specificity, immunogenicity, and versatility.
A break from evolution
Genome editors like CRISPR-Cas9 have powered a revolution in biotechnology and was awarded the 2020 Nobel Prize in Chemistry. This technology has enabled treatments like the first CRISPR-based cure for sickle cell disease (Casgevy), ex vivo engineered CAR-T cell therapies for cancer, and in vivo liver editing for transthyretin amyloidosis/
However, this system carries evolutionary baggage. Engineered for bacterial immunity, SpCas9 (the most widely used Cas protein in genome editing) tolerates off-target binding, triggers immune responses in humans, and is large enough to complicate delivery, all of which are non-desirable features.
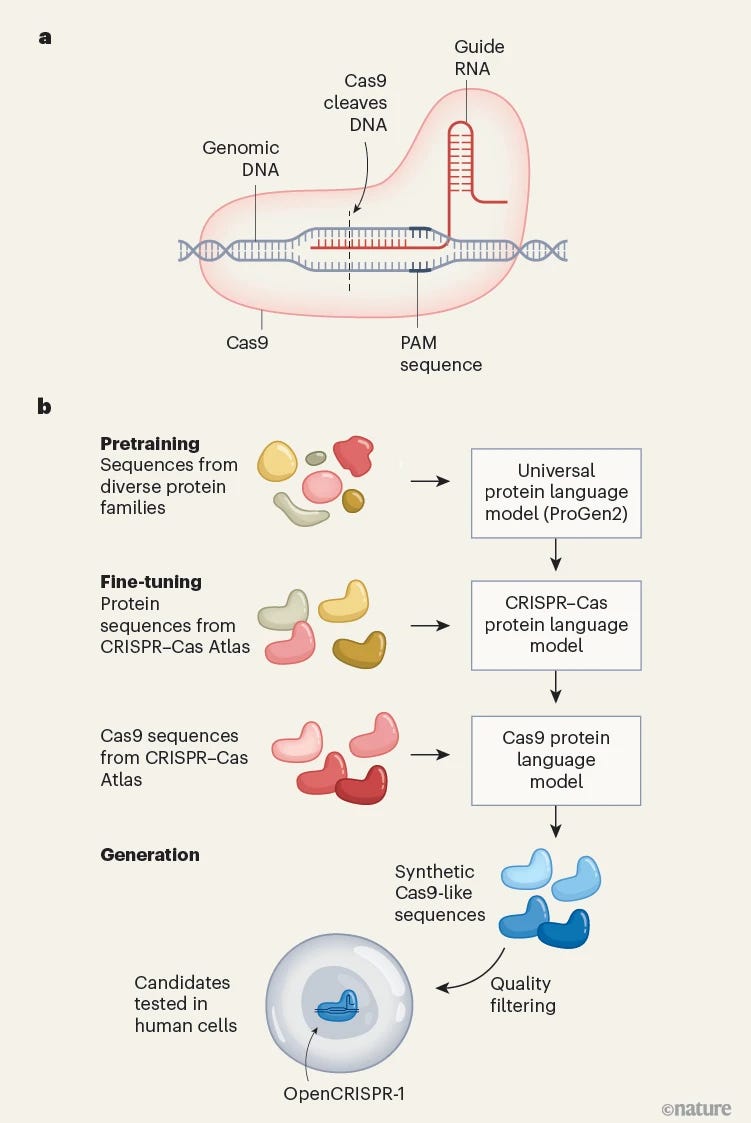
So how might we improve on this design? Learn from evolution and guide search to more optimal designs.
Profluent trained their large protein language model ProGen2 on a vast custom dataset called the CRISPR–Cas Atlas, mined from 26.2 terabases of microbial genomes. This yielded over 1.2 million CRISPR operons and more than 240,000 Cas9 sequences, offering an unparalleled training ground for AI to learn the statistical “grammar” of functional genome editors .
The power of generation
The model, fine-tuned on Cas-specific sequences, was prompted to generate 350,000 synthetic proteins, which were then filtered for quality and CRISPR compatibility. From these, 209 candidates were tested in human cells. The standout was OpenCRISPR-1, a protein 403 mutations away from SpCas9 and 180 mutations from its nearest natural cousin.
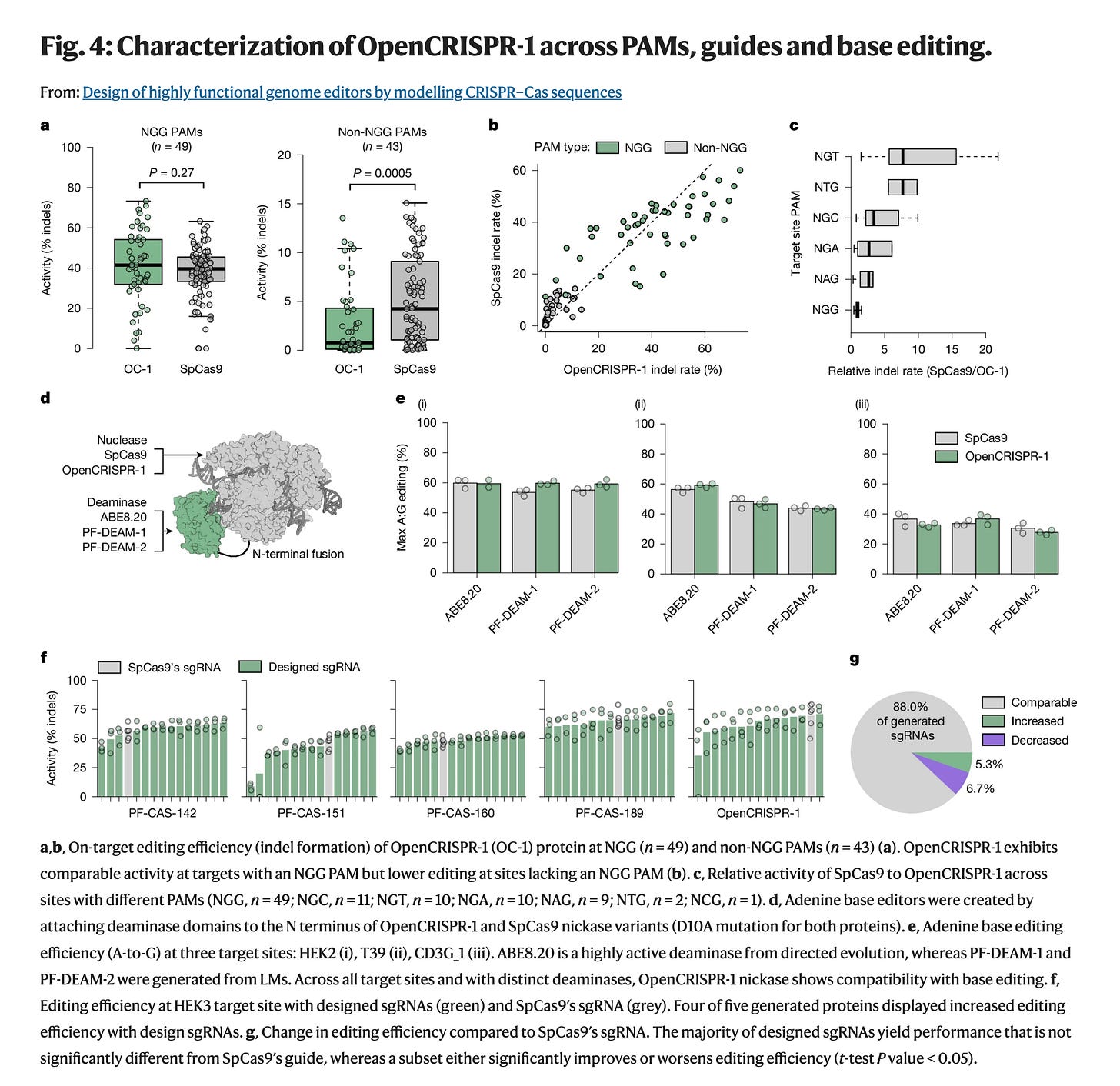
Despite this evolutionary sequence difference, it matched SpCas9’s on-target activity while reducing off-target edits by 95%!
Notably, OpenCRISPR-1’s off-target activity was a subset of SpCas9’s, with no new unintended edits. This suggests the editor is not just better, but safer, a critical threshold for therapeutic use.
Built-in therapeutic advantages
Profluent didn’t stop at editing efficiency. They assessed OpenCRISPR-1’s immunogenicity, finding that it lacks known T cell epitopes that plague SpCas9. In antibody assays across 40 donors, OpenCRISPR-1 showed consistently lower immune reactivity.
Further, it’s compatible with base editing and performs robust A-to-G conversions when fused to either evolved or AI-generated adenine deaminases. This hints at a future where entire gene editors, from protein to guide to payload, are AI-designed, component by component.
A new CRISPR era begins
OpenCRISPR-1 is not merely a new tool. It’s the first fully synthetic genome editor, born not from directed evolution or structural tinkering, but from massive-scale data-driven learning. It reflects a shift in biotech: from bioprospecting to bioengineering, from natural history to generative design.
And Profluent has open-sourced it all.
Plasmids for OpenCRISPR-1 are now live on Addgene, the central plasmid hub. Code, models, and sequences are available on GitHub and Zenodo. It is, in every sense, open science for labs, companies, and researchers around the world to build on.
Why this matters
CRISPR’s first chapter was defined by discovery. The next will be defined by design.
With OpenCRISPR-1, Profluent has proven that AI can leapfrog biology’s constraints to create entirely new proteins that are safe, specific, and clinically promising. The impact is immense: bespoke editors for rare diseases, compact versions for viral delivery, and lower immunogenicity for in vivo therapies.
AI didn’t just optimize biology, it created a new branch of it.
We’re proud to support Profluent as they lead the way into the programmable protein era.