Biology has scaling laws too
On Profluent’s launch of ProGen3, a new family of protein language models trained on over 3.4B protein sequences.
Introduction
Regular readers of Air Street Press will know that I’m long AI for science. Back in 2019 I wrote about 6 opportunities for AI-first biology. At the time, computer vision-powered microscopy and molecular design were the hot themes. Five years later, we have a Nobel Prize for protein structure prediction and biological datasets at scales I couldn’t have imagined during my PhD just a decade ago.
Meanwhile, generative AI continues its rapid climb up the hierarchy of human cognitive tasks. Scaling laws turned model building into a roadmap, offering predictable returns on compute and data investment. That simple fact reshaped how top labs operate. First formalized by Baidu Research in 2017 and popularized in large language models (LLMs), these laws have expanded to study of post-training and more recently for inference-time compute too.

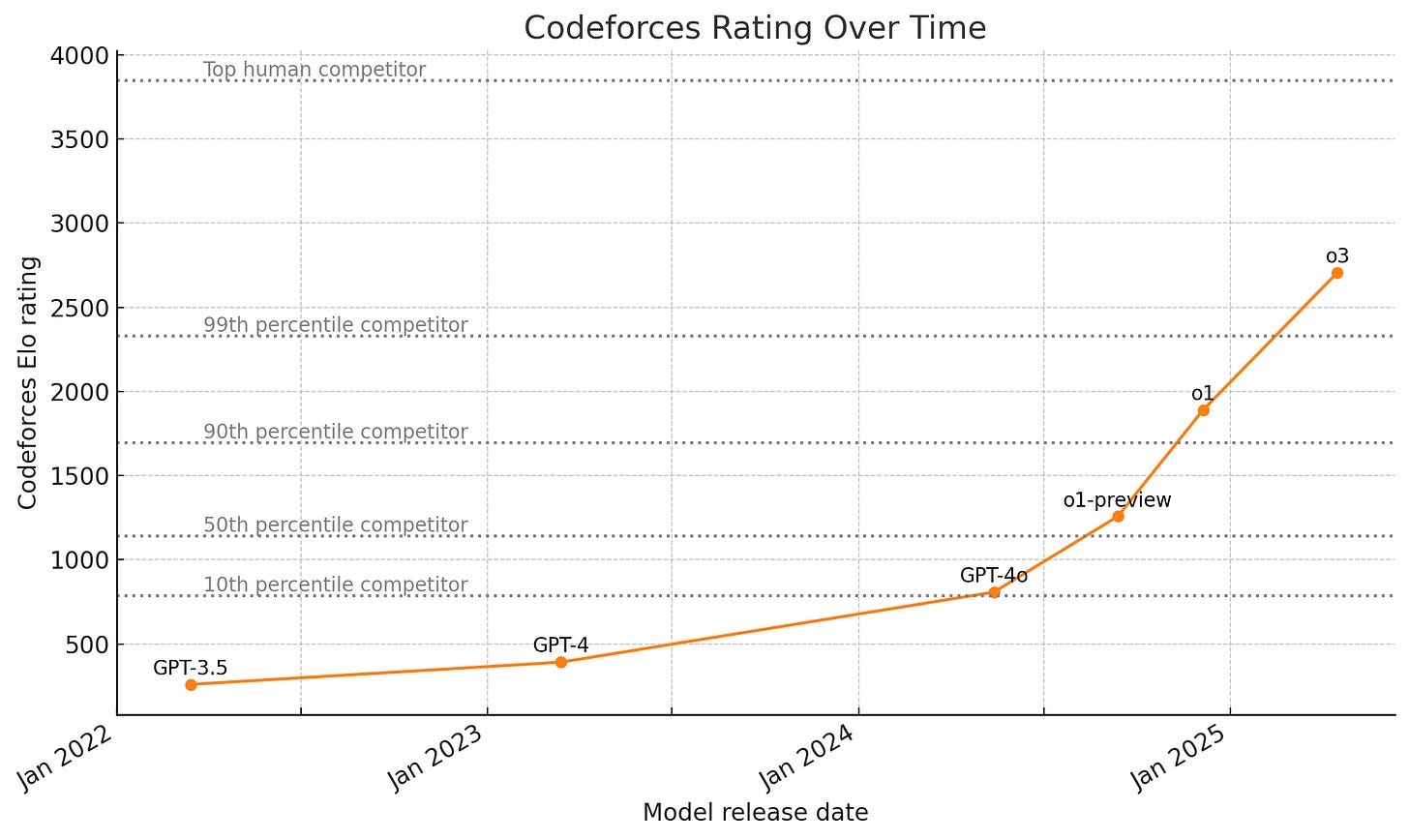
Indeed, reasoning models like o1 and o3 are starting to make real progress on domains like coding that were once considered far out of reach.
Scaling laws in biological design
So—what about biology? Let’s narrow in on proteins, where AI has had the largest impact so far. Natural evolution is a powerful search engine, but it’s slow—millions of years slow. To accelerate protein discovery, researchers have developed protein language models (PLMs) that can predict structure and function from raw amino acid sequences. These models, inspired by LLMs like GPT-2/3, treat biology as a sequence modeling problem.
ProGen, developed by the founding team of Profluent while at Salesforce Research and published in Nature Biotechnology in 2023, was an early example. It demonstrated that autoregressive models trained on protein sequences could generate novel, functional outputs across a wide range of families. ProGen2 expanded on this—but left open a set of important questions:
Do scaling laws hold in biology?
How should we optimize data distributions for training ever-larger PLMs?
Does scale improve the functional capabilities of generated proteins?
And do post-training alignment techniques work as well here as they do in human language?
Introducing ProGen3
To address these questions, Profluent have built ProGen3. It’s a family of generative PLMs built using a sparse mixture-of-experts architecture — similar to what powers state-of-the-art language models. The models are trained on the Profluent Protein Atlas v1, a rigorously curated dataset of 3.4B full-length proteins comprising 1.1T amino acid tokens.
This represents a 12x scale-up over previous models like ProGen2, and even exceeds the dataset used for Evolutionary Scale’s ESM-3. Model sizes range from 112M to 46B parameters, trained on 1.5T tokens, enabling both full sequence generation and infilling. Unlike most models, ProGen3’s output was also validated in the lab, showing real-world functionality, not just in silico benchmarks.
Crucially, ProGen3 isn’t just benchmarked in silico. The team synthesized a wide array of AI-generated proteins and tested them in wet-lab assays. The results: model outputs exhibit high fitness and functional diversity, with larger models benefiting most from alignment with lab data.
Scaling brings gifts
Profluent’s ProGen3 shows that scaling up the computational resources to pre-train PLMs predictably improves their validation losses for proteins both near and distant to their training data. The chart below shows how this result in proteins mirrors that in natural language from years prior:
So what more does scaling PLMs bring for protein engineers?
More valid sequences are generated, and repetition drops significantly.
Cluster diversity increases with larger models sampling from a wider and more diverse set of protein families.
Out-of-distribution generation improves as larger models access regions of sequence space unreachable by smaller ones.
Infilling quality rises, suggesting that larger models develop a deeper contextual understanding of protein constraints.
In vitro expression remains robust, even for sequences with just 30% similarity to known natural proteins.
Post-training alignment with lab data further boosts performance—for example, improving stability prediction—and larger models benefit more from alignment than smaller ones.
Next steps
ProGen3 is a compelling demonstration of how scale unlocks new capabilities in biological design. It shows that techniques developed in general-purpose AI—data curation, model scaling, sparse architectures, alignment—can directly inform progress in protein engineering.
The implications are broad: bespoke proteins for therapeutics, industrial enzymes, novel delivery vehicles, and beyond.
ProGen3 isn’t just a model, it’s proof that biology now follows its own scaling laws and we’re just starting to see what that unlocks.






